A team led by Prof. Zhen Yuan has successfully differentiated two distinct subtypes of prodromal Parkinson’s disease with the individualized brain radionics-based network, introducing insightful brain mechanisms in assisting clinical diagnosis and developing prevention strategies for individuals with prodromal Parkinson’s disease. The findings were published in a well-respected journal NeuroImage (5-year IF= 6.1, JCR Q1).
Parkinson’s disease can be divided into three stages based on the pathological changes, their degree of development, and clinical motor and non-motor symptoms: (1) Preclinical stage: only Parkinson’s disease pathological changes without any related symptoms; (2) Prodromal stage: non-motor symptoms or even mild motor symptoms appear, but do not meet the clinical diagnostic criteria for Parkinson’s disease. The risk of developing Parkinson’s disease within the next 10 years is extremely high; (3) Clinical stage: motor symptoms exist and meet the clinical diagnostic criteria.
Individuals in the prodromal phase of Parkinson’s disease (PD) exhibit significant heterogeneity and can be divided into distinct subtypes based on clinical symptoms, pathological mechanisms, and brain network patterns. However, no studies have been performed to inspect the subtyping of prodromal PD, which hinders its early diagnosis. Prof. Yuan’s lab tried to fill this research gap.
Prof. Yuan’s team constructed a radionics network for each individual from a large sample that included 110 normal controls (NC), 262 prodromal PD patients, and 108 PD patients. A nonlinear support vector machine classification algorithm was adopted to reveal the radiomics-based neural features significantly contributing to distinguishing between NC, prodromal PD, and PD patients, followed by a nonnegative matrix factorization method to cluster the subtypes of individuals with prodromal PD with 50 key radiomics-based connections (Fig 1). Such ingenious analyzing strategy can reliably identify two distinct subtypes of prodromal PD, including a subtype resembling the normal control group (N-P), individuals of which might be able to recover completely after treatment; the other subtype resembling the PD group (P-P), individuals of which can be targeted by intervention to delay the progression of symptoms.
Further analysis demonstrated these two subtypes of prodromal PD had distinct clinical assessment patterns (Fig 3 & Fig 4). Meanwhile, the multivariate approach of partial least squares analysis revealed the gene expression profiles were associated with the morphological changes in prodromal subtypes (Fig 5). Additional gene enrichment analyses also revealed the potential biological processes underlie the dissociation patterns of these two subtypes of prodromal PD (Fig 6).
This study offers valuable insights for the early, accurate diagnosis of PD and holds promise for the development of neuroprotective interventions tailored to individuals in the prodromal stage of the disease.
Original article link: https://doi.org/10.1016/j.neuroimage.2025.121012
Professor Zhen Yuan, Head of CCBS, is the corresponding author. Graduated PhD Dr. Lin Hua who now works at the Harvard Medical School is the first author. Graduated members from Prof. Yuan’s lab including Canpeng Huang, Dr. Xinglin Zeng, and Dr. Fei Gao also contributed to this study. This research is supported by the University of Macau (MYRG2022-00054-FHS, MYRG-GRG2023-00038-FHS-UMDF, and MYRG-GRG2024-00259-FHS), and the Macao Science and Technology Development Fund (FDCT 0014/2024/RIB1).
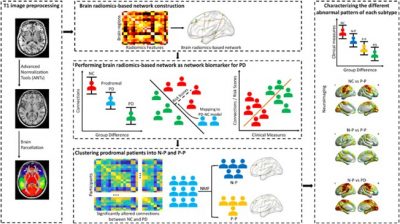
Figure 1 Procedures of clustering prodromal PD patients using individualized brain radiomics-based network
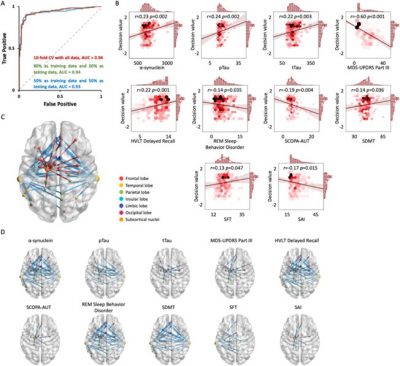
Figure 2 Classification performance between NC and PD based on brain radiomics-based network
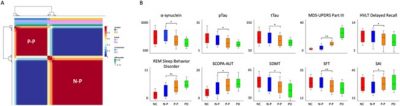
Fig. 3. Clustering results of the prodromal PD group and distinction of clinical measurements among NC, N-P, P-P, and PD groups
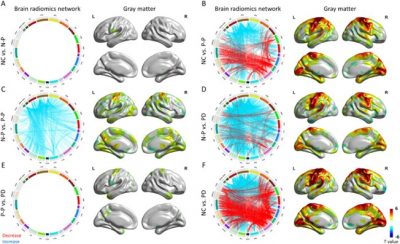
Fig. 4. Analysis of structural connectivity and morphometry among NC, N-P, P-P, and PD groups
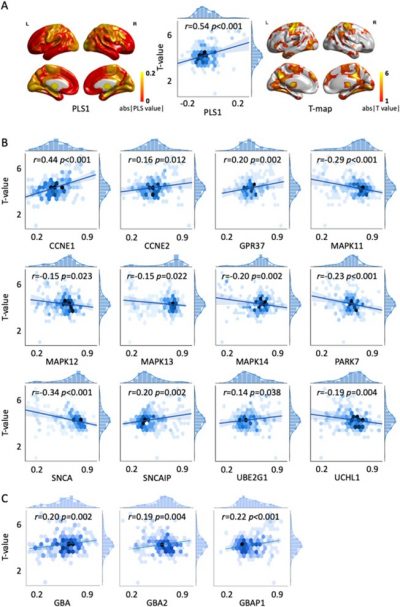
Fig. 5. Gene expressions were associated with GMV changes in prodromal subtypes

Fig. 6. Results of gene set enrichment analysis


